T013 · Data acquisition from PubChem¶
Note: This talktorial is a part of TeachOpenCADD, a platform that aims to teach domain-specific skills and to provide pipeline templates as starting points for research projects.
Authors:
Jaime Rodríguez-Guerra, 2019-2020, Volkamer lab, Charité
Dominique Sydow, 2019-2020, Volkamer lab, Charité
Yonghui Chen, 2019-2020, Volkamer lab, Charité
Aim of this talktorial¶
In this notebook, you will learn how to search for compounds similar to an input SMILES in PubChem with the API web service.
Contents in Theory¶
PubChem
Programmatic access to PubChem
Contents in Practical¶
Simple examples for the PubChem API
How to get the PubChem CID for a compound
Retrieve molecular properties based on a PubChem CID
Depict a compound with PubChem
Query PubChem for similar compounds
Determine a query compound
Create task and get the job key
Download results when job finished
Get canonical SMILES for resulting molecules
Show the results
References¶
Literature:
PubChem 2019 update: Nucleic Acids Res. (2019), 47, D1102-1109
PubChem in 2021: Nucleic Acids Res.(2021), 49, D1388–D1395
Documentation:
Theory¶
PubChem¶
PubChem is an open database containing chemical molecules and their measured activities against biological assays, maintained by the National Center for Biotechnology Information (NCBI), part of National Institutes of Health (NIH). It is the world’s largest freely available database of chemical information, collected from more than 770 data sources. There are three dynamically growing primary subdatabases, i.e., substances, compounds, and bioassay databases. As of August 2020, PubChem contains over 110 million unique chemical structures and over 270 million bioactivities (Nucleic Acids Res.(2021), 49, D1388–D1395).
These compounds can be queried using a broad range of properties including chemical structure, name, fragments, chemical formula, molecular weight, XlogP, hydrogen bond donor and acceptor count, etc. There is no doubt that PubChem has become a key chemical information resource for scientists, students, and the general public.
Every data from PubChem is free to access through both the web interface and a programmatic interface. Here, we are going to learn how to use the API of PubChem to do some cool things.
Programmatic access to PubChem¶
For some reasons, mainly historical, PubChem provides several ways of programmatic access to the open data.
PUG-REST is a Representational State Transfer (REST)-style version of the PUG (Power User Gateway) web service, with both the syntax of the HTTP requests and the available functions. It can also provide convenient access to information on PubChem records which are not reachable with other PUG services.
PUG-View is another REST-style web service for PubChem. It can provide full reports, including third-party textual annotation, for PubChem records.
PUG offers programmatic access to PubChem service via a single common gateway interface (CGI).
PUG-SOAP uses the simple object access protocol (SOAP) to access PubChem data.
PubChemRDF REST interface is a special interface for RDF-encoded PubChem data.
For more details about the PubChem API, please check out the introduction of programmatic access in the PubChem Docs.
In this tutorial, we will focus on the PUG-REST variant of the API, please refer to Talktorial T011 for an introduction.
Practical¶
In this section, we will discuss how to search PubChem based on a given query molecule using the PUG-REST access.
[1]:
import time
from pathlib import Path
from urllib.parse import quote
from IPython.display import Markdown, Image
import requests
import pandas as pd
from rdkit import Chem
from rdkit.Chem import PandasTools
from rdkit.Chem.Draw import MolsToGridImage
[2]:
HERE = Path(_dh[-1])
DATA = HERE / "data"
Simple examples for the PubChem API¶
Before querying PubChem for similar compounds, we provide some simple examples to show how to use the PubChem API. For more detail about the PubChem API, see PUG-REST tutorial.
How to get the PubChem CID for a compound¶
For example, we will search for the PubChem Compound Identification (CID) for Aspirin by name.
[3]:
# Get PubChem CID by name
name = "aspirin"
url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/{name}/cids/JSON"
r = requests.get(url)
r.raise_for_status()
response = r.json()
if "IdentifierList" in response:
cid = response["IdentifierList"]["CID"][0]
else:
raise ValueError(f"Could not find matches for compound: {name}")
print(f"PubChem CID for {name} is:\n{cid}")
# NBVAL_CHECK_OUTPUT
PubChem CID for aspirin is:
2244
Retrieve molecular properties based on a PubChem CID¶
We can get interesting properties for a compound through its PubChem CID, such as molecular weight, pKd, logP, etc. Here, we will search for the molecular weight for Aspirin.
[4]:
# Get mol weight for aspirin
url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/{cid}/property/MolecularWeight/JSON"
r = requests.get(url)
r.raise_for_status()
response = r.json()
if "PropertyTable" in response:
mol_weight = response["PropertyTable"]["Properties"][0]["MolecularWeight"]
else:
raise ValueError(f"Could not find matches for PubChem CID: {cid}")
print(f"Molecular weight for {name} is:\n{mol_weight}")
# NBVAL_CHECK_OUTPUT
Molecular weight for aspirin is:
180.16
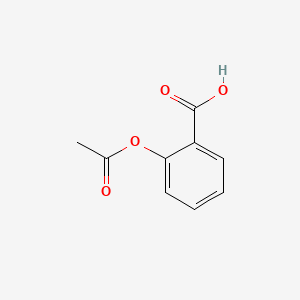
Depict a compound with PubChem¶
[5]:
# Get PNG image from PubChem
url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/{cid}/PNG"
r = requests.get(url)
r.raise_for_status()
display(Markdown("The 2D structure of Aspirin:"))
display(Image(r.content))
The 2D structure of Aspirin:
Query PubChem for similar compounds¶
Use a SMILES string of a query compound to search for similar compounds in PubChem.
Tip: You can check Talktorial T001 to see how to do a similar operation with the ChEMBL database.
We define a function to query the PubChem service for similar compounds to the given one. The Tanimoto-based similarity between (2D) PubChem fingerprint will be calculated here. You can see this paper for more details about similarity evaluation in PubChem, and the PubChem fingerprint specification for the details about PubChem fingerprint.
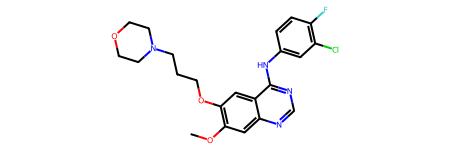
1. Determine a query compound¶
In the following steps, we will search compounds similar to Gefitinib, an inhibitor for EGFR, from the PubChem service. You can also choose another compound which you are interested in.
[6]:
query = "COC1=C(C=C2C(=C1)N=CN=C2NC3=CC(=C(C=C3)F)Cl)OCCCN4CCOCC4" # Gefitinib
print("The structure of Gefitinib:")
Chem.MolFromSmiles(query)
The structure of Gefitinib:
[6]:
2. Create task and get the job key¶
Input the canonical SMILES string for the given compound to create a new task, so we obtain a job key here. Note that this asynchronous API will not return the data immediately. In this case, the callback will be provided only when the requested resource is ready, which can be checked by the job key. So asynchronous requests are useful to work around certain slower operations.
[7]:
def query_pubchem_for_similar_compounds(smiles, threshold=75, n_records=10):
"""
Query PubChem for similar compounds and return the job key.
Parameters
----------
smiles : str
The canonical SMILES string for the given compound.
threshold : int
The threshold of similarity, default 75%. In PubChem, the default threshold is 90%.
n_records : int
The maximum number of feedback records.
Returns
-------
str
The job key from the PubChem web service.
"""
escaped_smiles = quote(smiles).replace("/", ".")
url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/similarity/smiles/{escaped_smiles}/JSON?Threshold={threshold}&MaxRecords={n_records}"
r = requests.get(url)
r.raise_for_status()
key = r.json()["Waiting"]["ListKey"]
return key
[8]:
job_key = query_pubchem_for_similar_compounds(query)
3. Download results when job finished¶
Check if the job finished within the time limit. The standard time limit in PubChem is 30 seconds (we can reset it). It means that if a request is not completed within the time limit, a timeout error will be returned. So, just wait patiently for the callback.
Then download results - PubChem CIDs in this case - when the job is finished.
[9]:
def check_and_download(key, attempts=30):
"""
Check job status and download PubChem CIDs when the job finished
Parameters
----------
key : str
The job key of the PubChem service.
attempts : int
The time waiting for the feedback from the PubChem service.
Returns
-------
list
The PubChem CIDs of similar compounds.
"""
url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/listkey/{key}/cids/JSON"
print(f"Querying for job {key} at URL {url}...", end="")
while attempts:
r = requests.get(url)
r.raise_for_status()
response = r.json()
if "IdentifierList" in response:
cids = response["IdentifierList"]["CID"]
break
attempts -= 1
print(".", end="")
time.sleep(10)
else:
raise ValueError(f"Could not find matches for job key: {key}")
return cids
[10]:
similar_cids = check_and_download(job_key)
Querying for job 624510668210559704 at URL https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/listkey/624510668210559704/cids/JSON...
4. Get canonical SMILES for resulting molecules¶
[11]:
def smiles_from_pubchem_cids(cids):
"""
Get the canonical SMILES string from the PubChem CIDs.
Parameters
----------
cids : list
A list of PubChem CIDs.
Returns
-------
list
The canonical SMILES strings of the PubChem CIDs.
"""
url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/{','.join(map(str, cids))}/property/CanonicalSMILES/JSON"
r = requests.get(url)
r.raise_for_status()
return [item["CanonicalSMILES"] for item in r.json()["PropertyTable"]["Properties"]]
[12]:
similar_smiles = smiles_from_pubchem_cids(similar_cids)
Then, we create the RDKit molecules and depict them.
[13]:
query_results_df = pd.DataFrame({"smiles": similar_smiles, "CIDs": similar_cids})
PandasTools.AddMoleculeColumnToFrame(query_results_df, smilesCol="smiles")
query_results_df.head(5)
[13]:
| smiles | CIDs | ROMol | |
|---|---|---|---|
| 0 | CN1CCC(CC1)COC2=C(C=C3C(=C2)N=CN=C3NC4=C(C=C(C... | 3081361 | <rdkit.Chem.rdchem.Mol object at 0x7f964f7dbe20> |
| 1 | CS(=O)(=O)CCNCC1=CC=C(O1)C2=CC3=C(C=C2)N=CN=C3... | 208908 | <rdkit.Chem.rdchem.Mol object at 0x7f964f7dbdc0> |
| 2 | COCCOC1=C(C=C2C(=C1)C(=NC=N2)NC3=CC=CC(=C3)C#C... | 176870 | <rdkit.Chem.rdchem.Mol object at 0x7f964f7dbe80> |
| 3 | COC1=C(C=C2C(=C1)C(=NC(=N2)N3CCN(CC3)C(=O)C4=C... | 68546 | <rdkit.Chem.rdchem.Mol object at 0x7f964f7dbee0> |
| 4 | CN(CCCNC(=O)C1CCCO1)C2=NC3=CC(=C(C=C3C(=N2)N)O... | 2092 | <rdkit.Chem.rdchem.Mol object at 0x7f964f7dbf40> |
5. Show the results¶
Show the compounds with images using RDKit’s drawing functions.
[14]:
def multi_preview_smiles(query_smiles, query_name, similar_molecules_pd):
"""
Show query and similar compounds in 2D structure representation.
Parameters
----------
query_smiles : str
The SMILES string of query compound.
query_name : str
The name of query compound.
similar_molecules_pd : pandas
The pandas DataFrame which contains the SMILES string and CIDs of similar molecules.
Returns
-------
MolsToGridImage
"""
legends = [f"PubChem CID: {str(s)}" for s in similar_molecules_pd["CIDs"].tolist()]
molecules = [Chem.MolFromSmiles(s) for s in similar_molecules_pd["smiles"]]
query_smiles = Chem.MolFromSmiles(query_smiles)
return MolsToGridImage(
[query_smiles] + molecules,
molsPerRow=3,
subImgSize=(300, 300),
maxMols=len(molecules),
legends=([query_name] + legends),
useSVG=True,
)
[15]:
print("The results of querying similar compounds for Gefitinib:")
img = multi_preview_smiles(query, "Gefitinib", query_results_df)
img
The results of querying similar compounds for Gefitinib:
~/.miniconda3/envs/teachopencadd/lib/python3.9/site-packages/rdkit/Chem/Draw/IPythonConsole.py:258: UserWarning: Truncating the list of molecules to be displayed to 10. Change the maxMols value to display more.
warnings.warn(
[15]:
Discussion¶
In this notebook, you have learned how to access and search similar compounds from the PubChem database via the PUG-REST programmatic access. Is it convenient? PUG-REST can do more than that. See PUG-REST to get more power!
Quiz¶
Can you make the similarity search more strict?
Is any of the proposed candidates already an approved inhibitor? (Hint: You can scrape PKIDB and check against the list of SMILES, also see Talktorial T011)
Can you try to reuse the functions in this notebook so search for similar compounds to Imatinib and inspect your results?